Bundschuh Research Group Projects
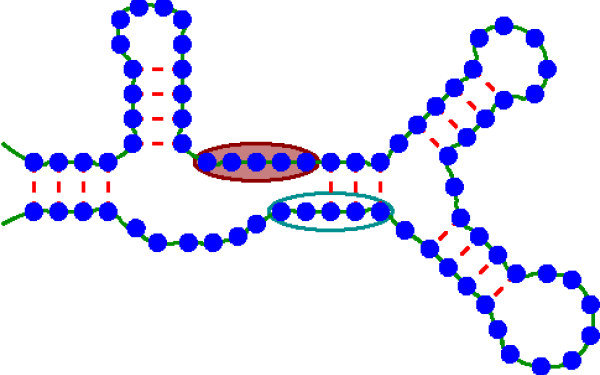
RNA Protein Interactions
RNA binding proteins (RBPs) affect every aspect of the life cycle of an RNA molecule including its processing, localization, stability, and translation efficiency.
It is thus important to understand which RBP binds which RNA where and how strongly.
One complication in answering these questions is that RNA molecules consist of nucleotides which like to form base pairs.
Depending on the RBP, these base pairings either suppress or favor RBP binding.
In either case, the binding energies of RBPs are on the same order of magnitude as the base pairing free energies, which implies that RBP binding and base pairing have to be modeled together rather than separately.
Thus, our lab has a long history of developing quantitative models of RNA-RBP interactions that take into account base pairing.
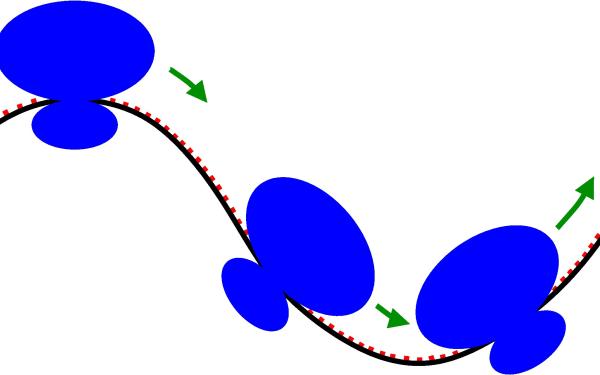
Prokaryotic Translation
Translation, the process of ribosomes "walking" along a messenger RNA, reading its information, and synthesizing a new protein based on this information is one of the most fundamental processes in Biology; every natural protein is made this way.
Specifically for prokaryotes (bacteria and archaea), understanding this process mechanistically is not only a question of curiosity about this fundamental process, but also of practical importance since a sizeable fraction of all antibiotics act on prokaryotic translation.
In collaboration with the Fredrick lab at OSU's Department of Microbiology, we develop and use multiple Genomics tools to elucidate the mechanism of prokaryotic translation.
On the one hand we take advantage of thousands of published and annotated prokaryotic genomes to compare and contrast how translational mechanisms vary across organisms.
On the other hand, we take advantage of next generation sequencing, specifically RNA-seq and ribosome sequencing, to analyze "snapshots" of the locations of millions of ribosomes simultaneously in prokaryotic cells under various conditions to learn what the various factors involved in prokaryotic translation are doing.

RNA Protein Interactions
RNA binding proteins (RBPs) affect every aspect of the life cycle of an RNA molecule including its processing, localization, stability, and translation efficiency.
It is thus important to understand which RBP binds which RNA where and how strongly.
One complication in answering these questions is that RNA molecules consist of nucleotides which like to form base pairs.
Depending on the RBP, these base pairings either suppress or favor RBP binding.
In either case, the binding energies of RBPs are on the same order of magnitude as the base pairing free energies, which implies that RBP binding and base pairing have to be modeled together rather than separately.
Thus, our lab has a long history of developing quantitative models of RNA-RBP interactions that take into account base pairing.

Prokaryotic Translation
Translation, the process of ribosomes "walking" along a messenger RNA, reading its information, and synthesizing a new protein based on this information is one of the most fundamental processes in Biology; every natural protein is made this way.
Specifically for prokaryotes (bacteria and archaea), understanding this process mechanistically is not only a question of curiosity about this fundamental process, but also of practical importance since a sizeable fraction of all antibiotics act on prokaryotic translation.
In collaboration with the Fredrick lab at OSU's Department of Microbiology, we develop and use multiple Genomics tools to elucidate the mechanism of prokaryotic translation.
On the one hand we take advantage of thousands of published and annotated prokaryotic genomes to compare and contrast how translational mechanisms vary across organisms.
On the other hand, we take advantage of next generation sequencing, specifically RNA-seq and ribosome sequencing, to analyze "snapshots" of the locations of millions of ribosomes simultaneously in prokaryotic cells under various conditions to learn what the various factors involved in prokaryotic translation are doing.
